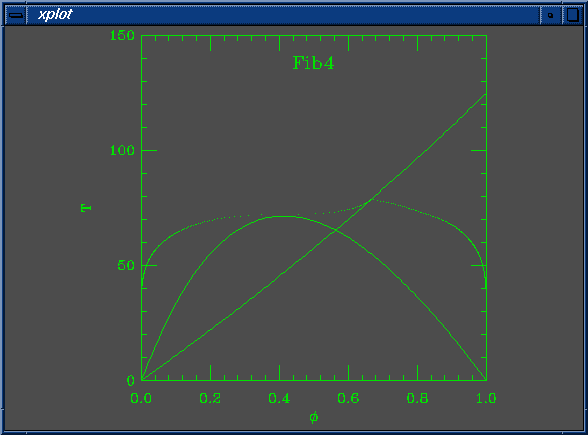
Here are a few phase diagrams, made when looking for a good set of parameters for which to find fibrils. We want Na > Nb, a reasonably large pure nematic region, and a spinodal that's not too far from the coexistence curve so that we can get phase separation without having to quench too far into the two phase region from the nematic. The diagrams are peculiar...
I'm including the relevant bits of the input files for the "catastrophe" program, so that you can see the parameters.
The solid lines in the curve are the Flory-Huggins spinodal and the nematic-isotropic transition line (B(phi) = W). The dots are coexistence points.
Jump to:

set Na = 200 set Nb = 100 set W0 = 5.490196e-03 set W1 = 4.000000e+00 set chi0 = 5.882353e-04 set chi1 = 1.000000e+00 set mustep = 1.000000e-03 set phimin = 1.000000e-03 set phimax = 1.000000e+00 set dphimax = 1.000000e-03
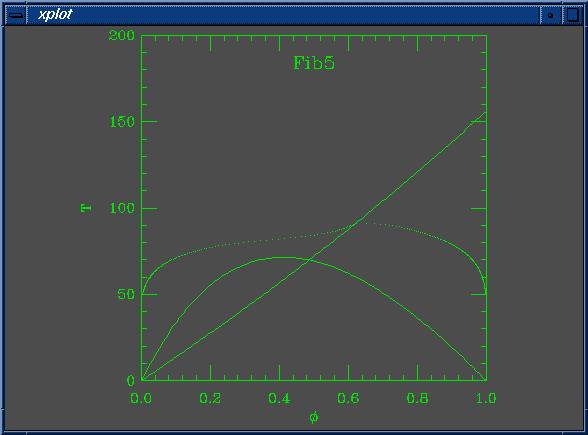
set Na = 200 set Nb = 100 set W0 = 5.490196e-03 set W1 = 5.000000e+00 set chi0 = 5.882353e-04 set chi1 = 1.000000e+00 set mustep = 1.000000e-02 set phimin = 1.000000e-03 set phimax = 1.000000e+00 set dphimax = 1.000000e-03
The spinodal doesn't touch the coexistence curve at the critical point!
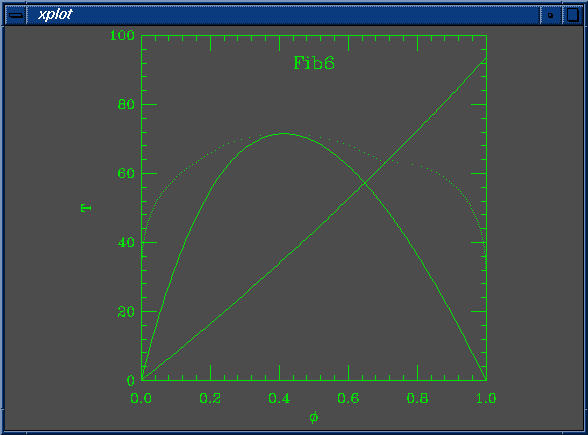
set Na = 200 set Nb = 100 set W0 = 5.490196e-03 set W1 = 3.000000e+00 set chi0 = 5.882353e-04 set chi1 = 1.000000e+00 set mustep = 1.000000e-03 set phimin = 1.000000e-04 set phimax = 9.999900e-01 set dphimax = 1.000000e-03

set Na = 200 set Nb = 100 set W0 = 3.000000e-02 set W1 = 1.000000e+00 set chi0 = 5.882353e-04 set chi1 = 1.000000e+00 set mustep = 1.000000e-02 set phimin = 1.000000e-03 set phimax = 1.000000e+00 set dphimax = 1.000000e-03
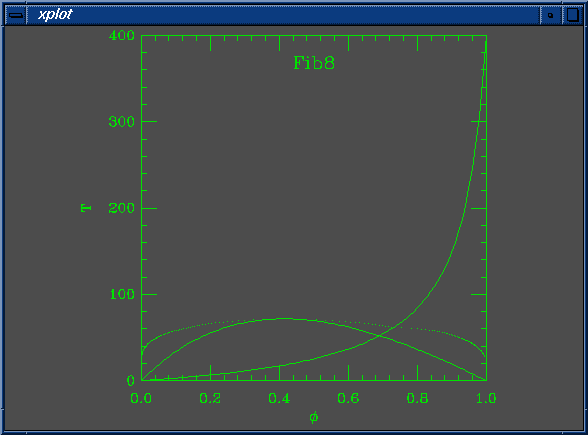
set Na = 200 set Nb = 100 set W0 = 3.500000e-02 set W1 = 1.000000e+00 set chi0 = 5.882353e-04 set chi1 = 1.000000e+00 set mustep = 1.000000e-02 set phimin = 1.000000e-03 set phimax = 1.000000e+00 set dphimax = 1.000000e-03
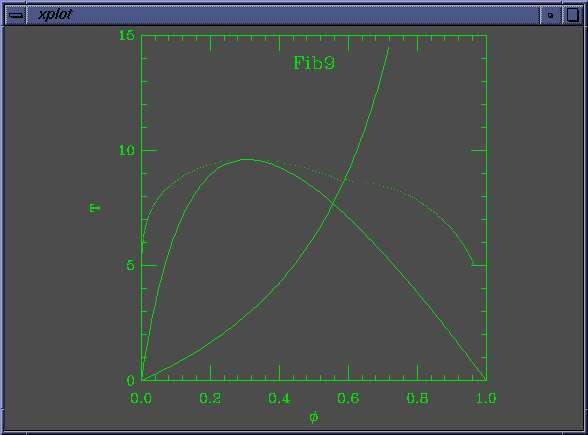
set Na = 50 set Nb = 10 set W0 = 1.400000e-01 set W1 = 1.000000e+00 set chi0 = 5.882353e-04 set chi1 = 1.000000e+00 set mustep = 1.000000e-01 set phimin = 1.000000e-04 set phimax = 9.999000e-01 set dphimax = 1.000000e-03
The NI transition is at T=100.
Here are some pictures of the time evolution of a quench to T=8, starting at phi=0.2, s=0. Here are some pictures of the evolution of an isolated droplet extracted from the later stages of this run.
Here are some pictures of another quench at T=8, phi=0.2, but starting with non-zero s, aligned in the x direction. This run has an interesting droplet merger.
Here are some pictures of a quench at T=6.5, starting at phi=0.6, s=0.1, theta=3 pi/10, and here are some more pictures of a run with the same physical parameters, but with higher spatial resolution. This run exhibits early stage fibrillar structures!
Starting from a fibrillar early stage of the previous run, and changing Nb and Gamma-phi-phi to simulate polymerization gives some later stage fibrillar structures. However, the same structures can be found by starting with an oriented unseparated configuration. Starting with a completely random configuration, however, gives a much less structured result.

set Na = 200 set Nb = 10 set W0 = 3.500000e-02 set W1 = 5.000000e-02 set chi0 = 5.882353e-04 set chi1 = 1.000000e+00 set mustep = 5.000000e-02 set phimin = 1.000000e-04 set phimax = 9.999000e-01 set dphimax = 1.000000e-03
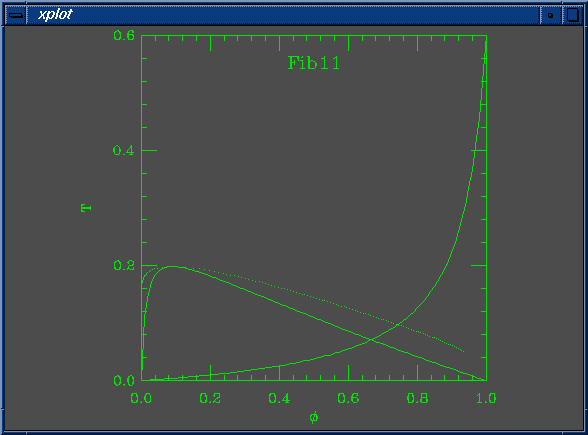
set Na = 100 set Nb = 1 set W0 = 7.000000e-02 set W1 = 3.000000e-03 set chi0 = 1.000000e-01 set chi1 = 1.000000e-01 set mustep = 1.000000e-03 set phimin = 1.000000e-04 set phimax = 9.999000e-01 set dphimax = 1.000000e-03
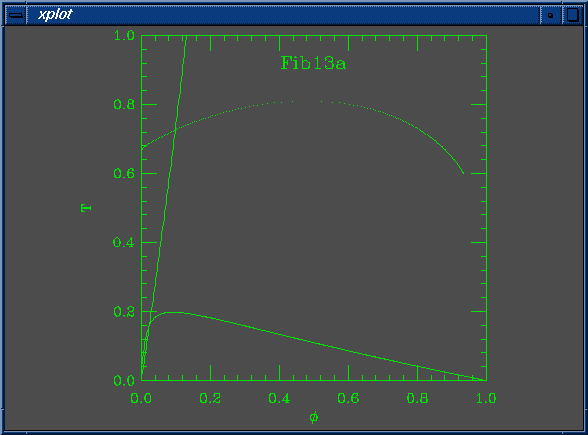
set Na = 100 set Nb = 1 set W0 = 7.400000e-02 set W1 = 5.000000e-01 set chi0 = 1.000000e-01 set chi1 = 1.000000e-01 set mustep = 1.000000e-03 set phimin = 1.000000e-04 set phimax = 9.999000e-01 set dphimax = 1.000000e-03

set Na = 100 set Nb = 1 set W0 = 7.400000e-02 set W1 = 2.500000e-01 set chi0 = 1.000000e-01 set chi1 = 1.000000e-01 set mustep = 1.000000e-03 set phimin = 1.000000e-200 set phimax = 1.000000e+00 set dphimax = 1.000000e-03