- Oxs_Exchange6Ngbr:
- Standard 6-neighbor exchange energy. The
exchange energy density contribution from cell i is given by
Ei = 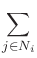 Aij Aij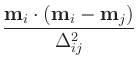
|
(7.1) |
where Ni is the set consisting of the 6 cells nearest to cell i,
Aij is the exchange coefficient between cells i and j in J/m,
and
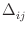 is the discretization step size between cell i and
cell j (in meters).
is the discretization step size between cell i and
cell j (in meters).
The Specify block for this term has the form
-
Specify Oxs_Exchange6Ngbr:name {
- default_A value
- atlas atlas_spec
- A {
-
region-1
region-1 A11
-
region-1
region-2 A12
- ...
-
region-m
region-n Amn
}
- }
or
-
Specify Oxs_Exchange6Ngbr:name {
- default_lex value
- atlas atlas_spec
- lex {
-
region-1
region-1 lex11
-
region-1
region-2 lex12
- ...
-
region-m
region-n lexmn
}
- }
where lex specifies the magnetostatic-exchange length, in
meters, defined by
lex = 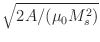 .
.
In the first case, the A block specifies Aij values
on a region by region basis, where the regions are labels declared by
atlas_spec. This allows for specification of A both
inside a given region (e.g., Aii) and along interfaces between
regions (e.g., Aij). By symmetry, if Aij is specified,
then the same value is automatically assigned to Aji as well.
The default_A value is applied to any otherwise
unassigned Aij.
In the second case, one specifies the magnetostatic-exchange length
instead of A, but the interpretation is otherwise analogous.
Although one may specify Aij (resp.
lexij) for any
pair of regions i and j, it is only required and only active if
the region pair are in contact. If long-range exchange interaction
is required, use Oxs_TwoSurfaceExchange.
In addition to the standard energy and field outputs,
Oxs_Exchange6Ngbr provides three scalar outputs involving the
angle between spins at neighboring cells:
- Max Spin Ang: maximum angle, in degrees, between
neigboring spins for the current magnetization state.
- Stage Max Spin Ang: Maximum value of Max Spin Ang
for the current stage.
- Run Max Spin Ang: Maximum value obtained by
Max Spin Ang during the simulation.
Examples: grill.mif, spinvalve.mif, tclshapes.mif.
- Oxs_UniformExchange:
- Similar to Oxs_Exchange6Ngbr, except the exchange constant A
(or exchange length lex) is uniform across all space. The
Specify block is very simple, consisting of either the label
A with the desired exchange coefficient value in J/m, or
the label lex with the desired magnetostatic-exchange
length in meters. Since A (resp. lex) is not spatially
varying, it is initialized with a simple constant (as opposed to a
scalar field object).
In addition to the standard energy and field outputs,
Oxs_UniformExchange provides the three scalar outputs
Max Spin Ang, Stage Max Spin Ang, and
Run Max Spin Ang as described for Oxs_Exchange6Ngbr.
These values are also accessible through the MIF GetStateData command.
Examples: sample.mif, cgtest.mif, stdprob3.mif.
- Oxs_ExchangePtwise:
- The exchange coefficient Ai is specified on
a point-by-point (or cell-by-cell) basis, as opposed to the pairwise
specification model used by Oxs_Exchange6Ngbr. The exchange
energy density at a cell i is computed across its nearest 6 neighbors,
Ni, using the formula
Ei = 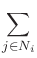 Aij, eff
Aij, eff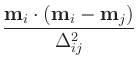
where
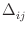 is the discretization step size from cell i to
cell j in meters, and
is the discretization step size from cell i to
cell j in meters, and
Aij, eff = 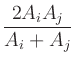 ,
,
with
Aij, eff = 0 if Ai and Aj are 0.
Note that
Aij, eff satisfies
the following properties:
| Aij, eff |
= |
Aji, eff |
|
| Aij, eff |
= |
Ai 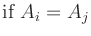 |
|
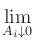 Aij, eff Aij, eff |
= |
0. |
|
Additionally, if Ai and Aj are non-negative,
min(Ai, Aj) 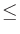 Aij, eff
Aij, eff 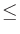 max(Ai, Aj).
max(Ai, Aj).
Evaluating the exchange energy with this formulation of
Aij, eff is equivalent to finding the minimum possible exchange energy
between cells i and j under the assumption that Ai and Aj
are constant in each of the two cells. Similar considerations are
made in computing the exchange energy for a 2D variable
thickness model [24].
The Specify block for Oxs_ExchangePtwise has the form
-
Specify Oxs_ExchangePtwise:name {
- A scalarfield_spec
- }
where scalarfield_spec is an arbitrary
scalar field object returning the desired exchange
coefficient in J/m.
In addition to the standard energy and field outputs,
Oxs_ExchangePtwise provides the three scalar outputs
Max Spin Ang, Stage Max Spin Ang, and
Run Max Spin Ang as described for Oxs_Exchange6Ngbr.
Example:
antidots-filled.mif.
- Oxs_TwoSurfaceExchange:
- Provides long-range bilinear and biquadratic exchange. Typically
used to simulate RKKY-style coupling across non-magnetic spacers in
spinvalves. The specify block has the form
-
Specify Oxs_TwoSurfaceExchange:name {
- sigma value
- sigma2 value
- surface1 {
- atlas atlas_spec
- region region_label
- scalarfield scalarfield_spec
- scalarvalue fieldvalue
- scalarside side
}
- surface2 {
- atlas atlas_spec
- region region_label
- scalarfield scalarfield_spec
- scalarvalue fieldvalue
- scalarside side
}
- }
Here sigma and sigma2 are the bilinear and
biquadratic surface (interfacial) exchange energies, in
J/m2. Either is optional, with default value 0.
The surface1 and surface2 sub-blocks describe
the two interacting surfaces. Each description consists of 5
name-values pairs, which must be listed in the order shown. In each
sub-block, atlas_spec specifies an atlas, and
region_label specifies a region in that atlas. These bound
the extent of the desired surface. The following
scalarfield, scalarvalue and
scalarside entries define a discretized surface inside the
bounding region. Here scalarfield_spec references a scalar
field object, fieldvalue should be a floating point value,
and side should be one of <, <=, >=, or
>. Any point for which the scalar field object takes a value
less than, less than or equal, greater than or equal, or greater
than, respectively, the scalarvalue value is considered to be
``inside'' the surface. (Values - and + for side
are deprecated synonyms for <= and >=.) The discretized
surface determined is the set of all points on the problem mesh that
are in the bounding region, are ``inside'' the surface, and have a
(nearest-) neighbor that is ``outside'' (i.e., not inside) the
surface. A neighbor is determined by the mesh; in a typical
rectangular mesh each cell has six neighbors.
In this way, 2 discrete lists of cells representing the two
surfaces are obtained. Each cell from the first list (representing
surface1) is then matched with the closest cell from the
second list (i.e., from surface2). Note the asymmetry in
this matching process: each cell from the first list is included in
exactly one match, but there may be cells in the second list that
are included in many match pairs, or in none. If the two surfaces
are of different sizes, then in practice typically the smaller will
be made the first surface, because this will usually lead to fewer
multiply-matched cells, but this designation is not required.
The resulting exchange energy density at cell i on one surface
from matching cell j on the other is given by
Eij = ![$\displaystyle {\frac{{\sigma\left[1-\textbf{m}_i\cdot\textbf{m}_j\right]
+\sig...
...\left[1-\left(\textbf{m}_i\cdot\textbf{m}_j\right)^2\right]
}}{{\Delta_{ij}}}}$](img16.gif)
where 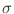 and
and 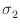 , respectively, are the bilinear and
biquadratic surface exchange coefficients between the two surfaces,
in J/m2,
mi and
mj are the normalized, unit spins
(i.e., magnetization directions) at cells i and j, and
, respectively, are the bilinear and
biquadratic surface exchange coefficients between the two surfaces,
in J/m2,
mi and
mj are the normalized, unit spins
(i.e., magnetization directions) at cells i and j, and
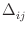 is the discretization cell size in the direction from
cell i towards cell j, in meters. Note that if
is the discretization cell size in the direction from
cell i towards cell j, in meters. Note that if 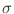 is
negative, then the surfaces will be anti-ferromagnetically coupled.
Likewise, if
is
negative, then the surfaces will be anti-ferromagnetically coupled.
Likewise, if 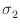 is negative, then the biquadratic term will
favor orthogonal alignment.
is negative, then the biquadratic term will
favor orthogonal alignment.
The following example produces an antiferromagnetic exchange coupling
between the lower surface of the ``top'' layer and the upper surface
of the ``bottom'' layer, across a middle ``spacer'' layer. The
simple Oxs_LinearScalarField object is used here to provide
level surfaces that are planes orthogonal to the z-axis. In
practice this example might represent a spinvalve, where the top and
bottom layers would be composed of ferromagnetic material and the
middle layer could be a copper spacer.
Specify Oxs_MultiAtlas:atlas {
atlas { Oxs_BoxAtlas {
name top
xrange {0 500e-9}
yrange {0 250e-9}
zrange {6e-9 9e-9}
} }
atlas { Oxs_BoxAtlas {
name spacer
xrange {0 500e-9}
yrange {0 250e-9}
zrange {3e-9 6e-9}
} }
atlas { Oxs_BoxAtlas {
name bottom
xrange {0 500e-9}
yrange {0 250e-9}
zrange {0 3e-9}
} }
}
Specify Oxs_LinearScalarField:zheight {
vector {0 0 1}
norm 1.0
}
Specify Oxs_TwoSurfaceExchange:AF {
sigma -1e-4
surface1 {
atlas :atlas
region bottom
scalarfield :zheight
scalarvalue 3e-9
scalarside <=
}
surface2 {
atlas :atlas
region top
scalarfield :zheight
scalarvalue 6e-9
scalarside >=
}
}
In addition to the standard energy and field outputs,
Oxs_TwoSurfaceExchange provides the three scalar outputs
Max Spin Ang, Stage Max Spin Ang, and
Run Max Spin Ang as described for Oxs_Exchange6Ngbr.
Example:
spinvalve-af.mif.
- Oxs_RandomSiteExchange:
- A randomized exchange energy. The Specify block has the form
-
Specify Oxs_RandomSiteExchange:name {
- linkprob probability
- Amin A_lower_bound
- Amax A_upper_bound
- }
Each adjacent pair of cells i, j, is given linkprob
probability of having a non-zero exchange coefficient Aij. Here
two cells are adjacent if they lie in each other's 6-neighborhood.
If a pair is found to have a non-zero exchange coefficient, then
Aij is drawn uniformly from the range
[Amin,Amax].
The exchange energy is computed using (7.1), the
formula used by the Oxs_Exchange6Ngbr energy object. The
value Aij for each pair of cells is determined during problem
initialization, and is held fixed thereafter. The limits
A_lower_bound and A_upper_bound may be any real
numbers; negative values may be used to weaken the exchange
interaction arising from other exchange energy terms. The only
restriction is that A_lower_bound must not be greater than
A_upper_bound. The linkprob value probability
must lie in the range [0, 1].
In addition to the standard energy and field outputs,
Oxs_RandomSiteExchange provides the three scalar outputs
Max Spin Ang, Stage Max Spin Ang, and
Run Max Spin Ang as described for Oxs_Exchange6Ngbr.
Example:
randexch.mif.
- Oxs_UZeeman:
- Uniform (homogeneous) applied field energy. This class is frequently
used for simulating hysteresis loops. The specify block takes an
optional multiplier entry, and a required field range list
Hrange. The field range list should be a compound list,
with each sublist consisting of 7 elements: the first 3 denote the
x, y, and z components of the start field for the range, the
next 3 denote the x, y, and z components of the end field for
the range, and the last element specifies the number of (linear) steps
through the range. If the step count is 0, then the range consists
of the start field only. If the step count is bigger than 0, then
the start field is skipped over if and only if it is the same field
that ended the previous range (if any).
The fields specified in the range entry are nominally in A/m, but
these values are multiplied by multiplier, which may be used to
effectively change the units. For example,
-
Specify Oxs_UZeeman {
- multiplier 795.77472
- Hrange {
- { 0 0 0 10 0 0 2 }
- { 10 0 0 0 0 0 1 }
}
- }
The applied field steps between 0 mT, 5 mT, 10 mT and back to 0 mT,
for four stages in total. If the first field in the second range
sublist was different from the second field in the first range
sublist, then a step would have been added between those field
values, so five stages would have resulted. In this example, note
that 795.77472=0.001/µ0.
In addition to the standard energy and field outputs, the
Oxs_UZeeman class provides these four scalar outputs:
- B: Magnitude of the applied field, in
mT. This is a non-negative quantity.
- Bx: Signed amplitude of the x-component
of the applied field, in mT.
- By: Signed amplitude of the y-component
of the applied field, in mT.
- Bz: Signed amplitude of the z-component
of the applied field, in mT.
Examples: sample.mif, cgtest.mif, marble.mif.
- Oxs_FixedZeeman:
- Non-uniform, non-time varying applied field.
This can be used to simulate a biasing field. The specify block
takes one required parameter, which defines the field, and one
optional parameter, which specifies a multiplication factor.
-
Specify Oxs_FixedZeeman:name {
- field vector_field_spec
- multiplier multiplier
- }
The default value for multiplier is 1. The field units,
after scaling by multiplier, should be A/m.
Examples: spinvalve.mif, spinvalve-af.mif, yoyo.mif.
- Oxs_ScriptUZeeman:
- Spatially uniform applied field,
potentially varying as a function of time and stage, determined by a
Tcl script. The Specify block has the form
-
Specify Oxs_ScriptUZeeman:name {
- script_args { args_request }
- script Tcl_script
- multiplier multiplier
- stage_count number_of_stages
- }
Here script indicates the Tcl script to use. The script
is called once each iteration. Appended to the script are the
arguments requested by script_args, in the manner
explained in the User Defined Support Procedures
section of the MIF 2 file format documentation. The value
args_request should be a subset of {stage
stage_time total_time base_state_id current_state_id}.
The units for the time options are seconds. The two
state_id options are intended for use with the
MIF GetStateData command;
refer to the documentation on that command in the MIF 2.1 section
for details. If script_args is not specified, the default
argument list is {stage stage_time total_time}.
The return value from the script should be a 6-tuple of numbers,
{Hx, Hy, Hz, dHx, dHy, dHz},
representing the applied field and the time derivative of the applied
field. The field as a function of time must be differentiable for
the duration of each stage. Discontinuities are permitted between
stages. If a time evolver is being used, then it is very important
that the time derivative values are correct; otherwise the evolver
will not function properly. This usual symptom of this problem is a
collapse in the time evolution step size.
The field and its time derivative are multiplied by the
multiplier value before use. The final field value should
be in A/m; if the Tcl script returns the field in T, then a
multiplier value of 1/µ0 (approx. 795774.72) should be
applied to convert the Tcl result into A/m. The default value for
multiplier is 1.
The stage_count parameter informs the
Oxs_Driver as to how many stages the
Oxs_ScriptUZeeman object wants. A value of 0 (the default)
indicates that the object is prepared for any range of stages. The
stage_count value given here must be compatible with the
stage_count setting in the driver Specify
block.
The following example produces a sinusoidally varying field of
frequency 1 GHz and amplitude 800 A/m, directed along the x-axis.
proc SineField { total_time } {
set PI [expr {4*atan(1.)}]
set Amp 800.0
set Freq [expr {1e9*(2*$PI)}]
set Hx [expr {$Amp*sin($Freq*$total_time)}]
set dHx [expr {$Amp*$Freq*cos($Freq*$total_time)}]
return [list $Hx 0 0 $dHx 0 0]
}
Specify Oxs_ScriptUZeeman {
script_args total_time
script SineField
}
In addition to the standard energy and field outputs, the
Oxs_ScriptUZeeman class provides these four scalar outputs:
- B: Magnitude of the applied field, in
mT. This is a non-negative quantity.
- Bx: Signed amplitude of the x-component
of the applied field, in mT.
- By: Signed amplitude of the y-component
of the applied field, in mT.
- Bz: Signed amplitude of the z-component
of the applied field, in mT.
Examples: acsample.mif, pulse.mif, rotate.mif,
varalpha.mif, yoyo.mif.
- Oxs_TransformZeeman:
- Essentially a combination of the Oxs_FixedZeeman and
Oxs_ScriptUZeeman classes, where an applied field is produced
by applying a spatially uniform, but time and stage varying linear
transform to a spatially varying but temporally static field. The
transform is specified by a Tcl script.
The Specify block has the form
-
Specify Oxs_TransformZeeman:name {
- field vector_field_spec
- type transform_type
- script Tcl_script
- script_args { args_request }
- multiplier multiplier
- stage_count number_of_stages
- }
The field specified by vector_field_spec is
evaluated during problem initialization and held throughout the life
of the problem. On each iteration, the specified Tcl script is called once. Appended to the script are the
arguments requested by script_args, as explained in the
User Defined Support Procedures section of the
MIF 2 file format documentation. The value for script_args
should be a subset of {stage stage_time total_time}.
The default value for script_args is the complete list in the
aforementioned order. The time arguments are specified in seconds.
The script return value should define a 3x3 linear transform and its
time derivative. The transform must be differentiable with respect
to time throughout each stage, but is allowed to be discontinuous
between stages. As noted in the Oxs_ScriptUZeeman
documentation, it is important that the derivative information be
correct. The transform is applied pointwise to the fixed
field obtained from vector_field_spec, which is
additionally scaled by multiplier. The
multiplier entry is optional, with default value 1.0.
The type transform_type value declares the
format of the result returned from the Tcl script. Recognized
formats are identity, diagonal, symmetric and
general. The most flexible is general, which indicates
that the return from the Tcl script is a list of 18 numbers,
defining a general 3x3 matrix and its 3x3 matrix of time derivatives.
The matrices are specified in row-major order, i.e., M1, 1,
M1, 2, M1, 3, M2, 1, M2, 2, .... Of course, this
is a long list to construct; if the desired transform is symmetric or
diagonal, then the type may be set accordingly to reduce the
size of the Tcl result string. Scripts of the symmetric type
return 12 numbers, the 6 upper diagonal entries in row-major order,
i.e., M1, 1, M1, 2, M1, 3, M2, 2, M2, 3,
M3, 3, for both the transformation matrix and its time
derivative. Use the diagonal type for diagonal matrices, in
which case the Tcl script result should be a list of 6 numbers.
The simplest transform_type is identity, which is the
default. This identifies the transform as the identity matrix, which
means that effectively no transform is applied, aside from the
multiplier option which is still active. For the identity
transform type, script and script_args should not be
specified, and Oxs_TransformZeeman becomes a clone of the
Oxs_FixedZeeman class.
The following example produces a 1000 A/m field that rotates in the
xy-plane at a frequency of 1 GHz:
proc Rotate { freq stage stagetime totaltime } {
global PI
set w [expr {$freq*2*$PI}]
set ct [expr {cos($w*$totaltime)}]
set mct [expr {-1*$ct}] ;# "mct" is "minus cosine (w)t"
set st [expr {sin($w*$totaltime)}]
set mst [expr {-1*$st}] ;# "mst" is "minus sine (w)t"
return [list $ct $mst 0 \
$st $ct 0 \
0 0 1 \
[expr {$w*$mst}] [expr {$w*$mct}] 0 \
[expr {$w*$ct}] [expr {$w*$mst}] 0 \
0 0 0]
}
Specify Oxs_TransformZeeman {
type general
script {Rotate 1e9}
field {0 1000. 0}
}
This particular effect could be obtained using the
Oxs_ScriptUZeeman class, because the field is uniform.
But the field was taken uniform only to simplify the example. The
vector_field_spec may be any Oxs vector field
object. For
example, the base field could be large in the center of the sample, and
decay towards the edges. In that case, the above example would generate
an applied rotating field that is concentrated in the center of the
sample.
The stage_count parameter informs the
Oxs_Driver as to how many stages the
Oxs_TransformZeeman object wants. A value of 0 (the default)
indicates that the object is prepared for any range of stages. The
stage_count value given here must be compatible with the
stage_count setting in the driver Specify
block.
Examples: sample2.mif, tickle.mif, rotatecenter.mif.
- Oxs_StageZeeman:
- The Oxs_StageZeeman class provides spatially varying applied
fields that are updated once per stage. In its general form, the
field at each stage is provided by an Oxs vector field
object
determined by a user supplied Tcl script. There is also a
simplified interface that accepts a list of vector
field files, one per
stage, that are used to specify the applied field.
The Specify block takes the form
-
Specify Oxs_StageZeeman:name {
- script Tcl_script
- files { list_of_files }
- stage_count number_of_stages
- multiplier multiplier
- }
The initialization string should specify either script or
files, but not both. If a script is specified,
then each time a new stage is started in the simulation, a Tcl command is formed by appending to Tcl_script the 0-based
integer stage number. This command should return a reference to an
Oxs_VectorField object, as either the instance name of an
object defined via a top-level Specify block elsewhere in the
MIF file, or as a two item list consisting of the name of an
Oxs_VectorField class and an appropriate initialization string.
In the latter case the Oxs_VectorField object will be created
as a temporary object via an inlined Specify call.
The following example should help clarify the use of the script
parameter.
proc SlidingField { xcutoff xrel yrel zrel } {
if {$xrel>$xcutoff} { return [list 0. 0. 0.] }
return [list 2e4 0. 0.]
}
proc SlidingFieldSpec { stage } {
set xcutoff [expr {double($stage)/10.}]
set spec Oxs_ScriptVectorField
lappend spec [subst {
atlas :atlas
script {SlidingField $xcutoff}
}]
return $spec
}
Specify Oxs_StageZeeman {
script SlidingFieldSpec
stage_count 11
}
The SlidingFieldSpec proc is used to generate the initialization
string for an Oxs_ScriptVectorField vector field object, which
in turn uses the SlidingField proc to specify the applied field
on a position-by-position basis. The resulting field will be
2e4 A/m in the positive x-direction at
all points with relative x-coordinate larger than $stage/10.,
and 0 otherwise. $stage is the stage index, which here is
one of 0, 1, ..., 10. For example, if $stage is 5, then the
left half of the sample will see a 2e4
A/m field directed to the right, and the right half of the sample
will see none. The return value from SlidingFieldSpec in this
case will be
Oxs_ScriptVectorField {
atlas :atlas
script {SlidingField 0.5}
}
The :atlas reference is to an Oxs_Atlas object defined
elsewhere in the MIF file.
The stage_count parameter lets the
Oxs_Driver know how many stages the
Oxs_StageZeeman object wants. A value of 0 indicates that the
object is prepared for any range of stages. Zero is the default
value for stage_count when using the Tcl_script
interface. The stage_count value given here must be compatible
with the stage_count setting in the driver
Specify block.
The example above made use of two scripts, one to specify the
Oxs_VectorField object, and one used internally by the
Oxs_ScriptVectorField object. But any Oxs_VectorField
class may be used, as in the next example.
proc FileField { stage } {
set filelist { field-a.ohf field-b.ohf field-c.ohf }
set spec Oxs_FileVectorField
lappend spec [subst {
atlas :atlas
file [lindex $filelist $stage]
}]
return $spec
}
Specify Oxs_StageZeeman {
script FileField
stage_count 3
}
The FileField proc yields a specification for an
Oxs_FileVectorField object that loads one of three files,
field-a.ohf, field-b.ohf, or field-c.ohf, depending on
the stage number.
Specifying applied fields from a sequence of files is common enough
to warrant a simplified interface. This is the purpose of the
files parameter:
Specify Oxs_StageZeeman {
files { field-a.ohf field-b.ohf field-c.ohf }
}
This is essentially equivalent to the preceding example, with two
differences. First, stage_count is not needed because
Oxs_StageZeeman knows the length of the list of files. You may
specify stage_count, but the default value is the length of the
files list. This is in contrast to the default value
of 0 when using the script interface. If stage_count is
set larger than the file list, then the last file is repeated as
necessary to reach the specified size.
The second difference is that no Oxs_Atlas is specified when
using the files interface. The Oxs_FileVectorField object
spatially scales the field read from the file to match a specified
volume. Typically a volume is specified by explicit reference to an
atlas, but with the files interface to Oxs_StageZeeman the
file fields are implicitly scaled to match the whole of the meshed
simulation volume. This is the most common case; to obtain a
different spatial scaling use the script interface as
illustrated above with a different atlas or an explicit x/y/z-range
specification.
The list_of_files value is interpreted as a
grouped list.
The remaining Oxs_StageZeeman parameter is
multiplier. The value of this parameter is applied as a
scale factor to the field magnitude on a point-by-point basis. For
example, if the field returned by the Oxs_VectorField object
were in Oe, instead of the required A/m, then multiplier could
be set to 79.5775 to perform the conversion. The direction of the
applied field can be reversed by supplying a negative multiplier
value.
In addition to the standard energy and field outputs, the
Oxs_StageZeeman class provides these four scalar outputs:
- B max: Pointwise maximum magnitude of the applied
field, in mT. This is a non-negative quantity;
B max = [(Bx max)2+(By max)2+(Bz max)2]1/2.
- Bx max: Signed value of the x-component of the applied
field at the point of maximum applied field magnitude, in mT.
- By max: Signed value of the y-component of the applied
field at the point of maximum applied field magnitude, in mT.
- Bz max: Signed value of the z-component of the applied
field at the point of maximum applied field magnitude, in mT.
Examples: sliding.mif, slidingproc.mif, rotatestage.mif,
rotatecenterstage.mif.
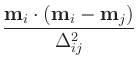
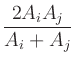 ,
,
![$\displaystyle {\frac{{\sigma\left[1-\textbf{m}_i\cdot\textbf{m}_j\right]
+\sig...
...\left[1-\left(\textbf{m}_i\cdot\textbf{m}_j\right)^2\right]
}}{{\Delta_{ij}}}}$](img16.gif)